Fibrosis in Crohn’s disease – what are the drivers in the mucosal microenvironment?
Prinicipal Investigator

Dr. Lea Maxie Haag
Scientific interest within the context of the graduate college:
Based at the Department of Gastroenterology, Infectious Diseases and Rheumatology at Campus Benjamin Franklin, Charité – Universitätsmedizin Berlin and at the Max Delbrück Center (MDC) in Berlin, our main clinical and research focus are chronic inflammatory diseases like Inflammatory bowel diseases (IBD). Crohn’s disease (CD), one of the main forms of IBD, has different clinical phenotypes, including an inflammatory (B1) as well as a stricturing (B2) disease phenotype, in which patients develop fibrotic stenoses of the intestine. Developing a molecular understanding of why some patients suffer from the formation of fibrotic strictures, while others do not, is still not understood. By investigating the mucosal microenvironment, we aim to establish a holistic understanding of the process of fibrotic stricture formation in CD to find answers to the driving question: ‘Stricturing in Crohn’s disease – what are the drivers in the mucosal microenvironment?’
Project description:
What is the underlying mechanism leading to the formation of fibrotic strictures in Crohn’s disease? Why do some patients develop a stricturing disease phenotype while others do not?
Within this project, we aim to better understand mechanisms responsible for the development of a stricturing phenotype. We hypothesize that the mucosal microenvironment in patients with an inflammatory CD phenotype (B1) differs from the microenvironment found in patients with a stricturing phenotype (B2). Therefore, the mandatory prerequisite is to deeply characterize the different disease phenotypes. The proposed project will be associated to the clinical study ‘InFlame’ (DRKS00031203), which is already running since early 2023. We have developed a ‘mucosa phenotyping pipeline’ (Figure 1) with high-throughput methods to analyze the intestinal microenvironment e.g., interstitial fluid and mucosal microbiota from biopsies, 3D histology and imaging mass cytometry as well as components of the peripheral immune system via blood samples, to comprehensively phenotype IBD patients and healthy individuals.
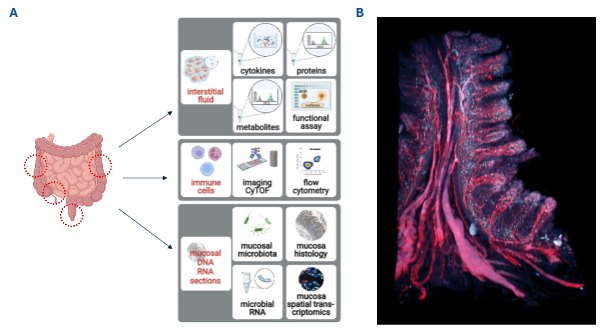
Figure 1. Mucosa Phenotyping pipeline: A. Biopsies will be obtained during endoscopy at the indicated red circles and handled as described in detail in the main text. B. Human intestinal biopsy stained in red with nanobodies against CD31 (vessels) and podoplanin (lymphatics) and blue against CD8 T cells.
All methods are established and running in the lab. The existing study team and the ongoing recruitment process guarantee intensive support and the provision of all necessary infrastructure.
WP1. Identification of Crohn’s Disease Inflammatory Subtypes. Identify Crohn’s disease patients already recruited who either have an inflammatory B1-phenotype or a stricturing B2-phenotype.
WP2. Deep phenotyping by our mucosa phenotyping platform. Biopsies will be used for (i) isolation of IF, (ii) phenotyping of the immune- cells including 3D histology (iii) analysis of mucosal microbiota. This approach will result in an in-depth spatiofunctional and spatiomechanistic atlas for microbes, metabolites, proteins, cytokines, immune cells, and transcriptomes from different segments of the intestinal tract.
References
- Avery EG*, Haag LM*, McParland V, Kedziora SM, Zigra GJ, Valdes DS, […], Siegmund B, Wiig H, Müller DN. Intestinal interstitial fluid isolation provides novel insight into the human host-microbiome interface. Cardiovasc Res. 2025; 121(5):803-816. *equal contribution
- Stankey CT*, Bourges C*, Haag LM*, Turner-Stokes T, Piedade AP, Palmer-Jones C, […], Wallace C, Thomas DC, Lee JC. A disease-associated gene desert directs macrophage inflammation through ETS2. Nature. 2024; 630(8016):447-456. *equal contribution
- Rieder F and Zimmermann EM. Crohn’s disease complicated by strictures: a systematic review. Gut. 2013; 62(7):1072-1084.
- Rieder F and Murkherjee PK. Fibrosis in IBD: from pathogenesis to therapeutic targets. Gut. 2024; 73(5):854-866.