Transgenerational effects of impaired metabolic adaptation during pregnancy
Principal Investigator
Scientific interest within the context of the graduate college:
We study development and function of the innate immune system, in particular of innate lymphoid cells (ILC). A current focus is to obtain a molecular understanding of how the innate immune system, by integrating environmental signals (such as those derived from nutrients, microbiota, circadian rhythm) contributes to tissue physiology. Recent studies have revealed ever more intriguing relationships between innate immune system components and basic developmental and biologic processes that are likely to reveal unsuspected pathways by which the immune system might be plumbed to improve health and healthspan. These lines of research have suggested new functions of the immune system for processes such as tissue homeostasis, morphogenesis, metabolism, regeneration and growth. Our research is developing by crossing boundaries of disciplines (immunology, microbiology, developmental biology, stem cell biology, tumor biology, regenerative medicine etc.) and is, by nature, highly interdisciplinary.
Project description:
Recent findings from our laboratory demonstrate that pregnancy and lactation are accompanied by significant intestinal growth in the mother. This physiological adaptation is essential to meet the increased metabolic demands associated with gestation and nursing. Importantly, we have identified that cues from the innate immune system are required to support this intestinal remodeling (manuscript in preparation).
In mouse models, impaired maternal intestinal growth and metabolic adaptation result in offspring with lower birth weights, which normalize within 2-3 weeks postpartum. However, these offspring exhibit a heightened susceptibility to inflammatory and metabolic diseases in adulthood. Using a TNF-driven model of inflammatory bowel disease (IBD), we observed that progeny of mothers with impaired intestinal adaptation have decreased frequencies of regulatory T cells (Tregs) and increased infiltration of pro-inflammatory neutrophils in affected tissues.
The mechanisms by which reduced nutrient availability during pregnancy and lactation predispose offspring to long-term inflammatory disorders are currently unknown. We hypothesize that impaired maternal metabolic adaptation leads to alterations in nutrient transfer, resulting in metabolic and epigenetic reprogramming of affected tissues (i.e., intestine) or the offspring’s immune system—particularly in primary immune organs such as the bone marrow and thymus.
In this proposal, we aim to define how impaired maternal intestinal and metabolic adaptation shapes the long-term immune landscape of the offspring.
Aim 1: Characterize the tissue architecture, transcriptional profiles, and epigenetic programs in offspring born to mothers with impaired intestinal growth and metabolic adaptation. We will use high-resolution clonal lineage tracing, single-cell transcriptomics, and epigenomic profiling to identify changes in key metabolic and immune tissues over time.
Aim 2: Establish causal links between maternal metabolic perturbation and immune regulatory dysfunction in offspring. Using genetic and metabolic interventions, we will test whether restoring maternal intestinal adaptation can reverse immune dysregulation in the offspring and explore potential avenues for therapeutic modulation of inflammatory susceptibility.
This project will define a previously unappreciated link between maternal metabolic adaptation and immune programming in offspring. The outcomes are expected to provide foundational knowledge for developing early-life interventions to prevent chronic inflammatory diseases.
References
- Biniaris-Georgallis SI, Aschman T, Stergioula K, Schreiber F, Jafari V, Taranko A, […], Diefenbach A*, Kanda M*, Triantafyllopoulou A*. Amplification of autoimmune organ damage by NKp46-activated ILC1s. Nature. 2024; 634(8035):952-960. *equally contributing senior authors
- Witkowski M, Tizian C, Ferreira-Gomes M, Niemeyer D, Jones TC, Heinrich F, […], Radbruch A, Mashreghi MF, Diefenbach A. Untimely TGFβ responses in severe COVID-19 limit antiviral function of NK cells. Nature. 2021; 600(7888):295-301.
- Guendel F, Kofoed-Branzk M, Gronke K, Tizian C, Witkowski M, Cheng HW, […], Mashreghi MF, Kruglov AA, Diefenbach A. Group 3 innate lymphoid cells program a distinct subset of IL-22BP-producing dendritic cells demarcating solitary intestinal lymphoid tissues. Immunity. 2020; 53(5):1015-1032.
- Schaupp L, Muth S, Rogell L, Kofoed-Branzk M, Melchior F, Lienenklaus S, […], Schild H, Diefenbach A1,*, Probst HC*. Microbiota-Induced Type I Interferons Instruct a Poised Basal State of Dendritic Cells. Cell. 2020; 181(5):1080-1096.e19. 1lead senior author; *equally contributing senior authors
- Gronke K, Hernández PP, Zimmermann J, Klose CSN, Kofoed-Branzk M, Guendel F, […], Glatt H, Triantafyllopoulou A, Diefenbach A. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature. 2019; 566(7743):249-253.
The role of the gut microbiome in mediating kidney disease associated comorbidities
Principal Investigator

Dr. Felix Behrens
Scientific interest within the context of the graduate college:
Chronic kidney disease is a major global health concern of increasing prevalence, driving a plethora of secondary comorbidities. A mechanistic understanding of the driving factors is essential to develop preventive and therapeutic concepts. Our research group investigates microbiome-mediated mechanisms of cardiovascular risk (e.g. Wilck et al. Nature 2017, Bartolomaeus et al. Circulation 2019, Avery et al. Cardiovascular Research 2023; see below for more). We believe that CKD is an understudied risk factor for cardiovascular disease, characterized by changes in microbiome composition. In the long term, microbiome-targeting interventions could help to reduce cardiovascular risk in CKD patients.
Project description:
Chronic kidney disease (CKD) is a main contributor to cardiovascular (CV) risk, inflammation and subsequent multimorbid conditions. CKD associates with dysbiotic alterations of the gut microbiome, a resulting dysbalance of gut bacterial metabolites, an impaired intestinal barrier function and resulting chronic systemic inflammation driving CV disease. We have recently shown that intestinal barrier dysfunction and increased AhR activity may drive inflammation and cardiovascular remodeling (Holle et al. JASN 2022; Holle et al. manuscript in preparation). The aim of the present project is to investigate the transmission of CKD-associated traits by microbiome transfer from humans to mice. Further biomaterials from human cohort materials will be used to understand if the human microbiome in mice can be compared to the “real world”.
Aim 1: Fecal microbiota transfer (FMT) of human microbiota into mice. We have prior experience in FMT experiments. As such, mice can be used as an experimental platform to understand the efficacy of microbiome-targeting interventions. We therefore use mice (colonized with microbiota from CKD patients and controls) to perform dietary interventions to improve microbiome dysbiosis and associated pathologies (gut barrier dysfunction, inflammation etc.). In the present experiment we aim to investigate the potential of dietary fiber to improve CKD-associated microbiome dysbiosis. We aim to analyze the microbiome by sequencing, the immune phenotype (flow cytometry) as well as cardiovascular alterations (e.g. cardiac hypertrophy and fibrosis by histology, gene expression). These initial data will inform further in-depth analyses.
Aim 2: Investigation of the influence of CKD candidate bacterial species on AhR and the intestinal barrier. Gnotobiotic mice, i.e. mice with known, defined gut bacteria, provide an opportunity to mechanistically study the impact of specific bacteria on the host organism. In previous microbiome analyses (Holle et al. JASN 2022; Holle et al. manuscript in preparation), we have identified CKD-specific candidate species that we would like to investigate by colonizing germ-free mice. The analysis will focus on the intestinal barrier (immunohistology, gene expression), aryl hydrocarbon receptor activity (cell-based reporter assay) and inflammation (flow cytometry from whole blood, spleen, intestine, heart and kidney).
Aim 3: Analysis of biosamples from a human CKD cohort. We conducted a fiber-intervention (placebo-controlled) in dialysis patients. We aim to analyze the microbiome, markers of a “leaky” gut as well as the immune phenotype (flow cytometry). The results will be compared to the data generated in mice.
References
- Holle J, Reitmeir R, Behrens F, Singh D, Schindler D, Potapenko O, […], Wilck N, Löber U, Bartolomaeus H. Gut microbiome alterations precede graft rejection in kidney transplantation patients. Am J Transplant. 2025; S1600-6135(25)00093-0. Online ahead of print.
- Wimmer MI, Bartolomaeus H, Anandakumar H, Chen CY, Vecera V, Kedziora S, […], Forslund-Startceva SK, Müller DN, Wilck N. Metformin modulates microbiota and improves blood pressure and cardiac remodeling in a rat model of hypertension. Acta Physiol (Oxf). 2024; 240(11):e14226.
- Holle J, McParland V, Anandakumar H, Gerritzmann F, Behrens F, Schumacher F, […], Kleuser B, Bartolomaeus H, Wilck N. Gut dysbiosis contributes to TMAO accumulation in CKD. Nephrol Dial Transplant. 2024; 39(11):1923-1926
- Dörner PJ, Anandakumar H, Röwekamp I, Fiocca Vernengo F, Millet Pascual-Leone B, Krzanowski M, […], Bartolomaeus H, Heimesaat MM, Opitz B. Clinically used broad-spectrum antibiotics compromise inflammatory monocyte-dependent antibacterial defense in the lung. Nat Commun. 2024; 15(1):2788.
- Anandakumar H, Rauch A, Wimmer MI, Yarritu A, Koch G, McParland V, Bartolomaeus H, Wilck N. Segmental patterning of microbiota and immune cells in the murine intestinal tract. Gut Microbes. 2024; 16(1):2398126.
- Avery EG, Bartolomaeus H, Rauch A, Chen CY, N’Diaye G, Lober U, […], Forslund SK, Muller DN, Wilck N. Quantifying the impact of gut microbiota on inflammation and hypertensive organ damage. Cardiovasc Res. 2023; 119(6):1441-1452.
- Holle J, Bartolomaeus H, Lober U, Behrens F, Bartolomaeus TUP, Anandakumar H, […], Kirwan JA, Wilck N, Muller D. Inflammation in Children with CKD Linked to Gut Dysbiosis and Metabolite Imbalance. J Am Soc Nephrol. 2022; 33(12):2259-2275.
- Maifeld A, Bartolomaeus H, Löber U, Avery EG, Steckhan N, Markó L, […], Michalsen A, Müller DN, Forslund SK.Fasting alters the gut microbiome reducing blood pressure and body weight in metabolic syndrome patients. Nat Commun. 2021; 12(1):1970.
- Bartolomaeus H, Balogh A, Yakoub M, Homann S, Marko L, Hoges S, […], Muller DN, Stegbauer J, Wilck N. Short-Chain Fatty Acid Propionate Protects From Hypertensive Cardiovascular Damage. Circulation. 2019; 139(11):1407-1421.
- Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, […], Linker RA, Alm EJ, Muller DN. Salt-responsive gut commensal modulates T(H)17 axis and disease. Nature. 2017; 551(7682):585-589.
Homeostatic mechanisms supporting the maintenance of tissue-resident macrophages
Principal Investigator
Scientific interest within the context of the graduate college:
Active maintenance of tissue health requires maintenance of tissue-resident macrophages that perform homeostatic functions. Loss of tissue-resident macrophages reduces the ability of tissues to maintain homeostasis or to return to homeostasis after inflammation. We propose that exploring the mechanisms that maintain tissue-resident macrophages will allow us to identify molecular targets that promote tissue health and to achieve deeper remission after treatment of chronic inflammatory diseases.
Project description:
Tissue-resident macrophages play an active role in maintaining tissue homeostasis. The clearance of apoptotic cells and other tissue debris, the immediate immune response to pathogens, and metabolic regulation are all functions of tissue resident macrophages that are essential for tissue health. Failure to maintain the tissue-resident macrophage population during aging, or loss of tissue-resident macrophages in chronic inflammation results in loss of tissue function. Despite their importance, the mechanisms that actively maintain tissue-resident macrophages in healthy tissues are largely unknown. The current project is based on extensive preliminary data from our lab that have uncovered a novel pathway required for the maintenance of tissue-resident macrophages.
The project will explore the mechanisms that maintain tissue-resident macrophages across different organs, including macrophage populations of different origins, and in response to various acute inflammatory stimuli, such as TNF and type I interferons. The project will further compare the mechanisms that regulate tissue-resident macrophage populations in homeostasis versus in chronic inflammatory diseases. Overall, this line of work aims to uncover essential pathways that promote tissue-resident macrophage maintenance and thus tissue health, paving the way for a deeper remission following treatment of chronic inflammatory diseases.
The human assembloid reveals the crosstalk of colonic resident macrophages with their neighboring epithelium, vasculature, and enteric nervous system
Principal Investigator
Scientific interest within the context of the graduate college:
Our lab investigates the gastrointestinal (GI) tract epithelium – the cellular layer lining the stomach and intestine that forms a vital barrier between the body and the external environment. A central focus lies on understanding the mechanisms of epithelial regeneration and how interactions with commensal microbiota and pathogenic bacteria influence this process, potentially leading to inflammation, gut dysfunction, or cancer.
Project description:
The crypt-villus architecture of the intestinal mucosa is underpinned by dynamic interactions between distinct populations of epithelial, stromal, and immune cells. While the epithelial compartment has garnered significant attention, there is a growing appreciation for the critical role of mesenchymal cells in shaping epithelial stem cell function and dictating lineage specification in homeostasis and upon perturbation.
We hypothesize that similar processes are co-opted in the context of carcinogenic transformation, that in contrast to the traditional view does not simply rely on mutational events but instead emerges as a result of a dynamic interplay between epithelium and stroma. Using a model of gastric cancrinogenesis that integrates mutational and environmental cues we aim to explore this process in detail and would like to integrate a doctoral thesis into this project.
Aim 1: To characterize the microenvironemnt in a murine model of gastric cancer.
Aim 2: To functionally determine key stromal drivers of epithelial transformation in the stomach.
Aim 3: To explore the relevance of the identified pathways in a cohort of patients with gastric cancer.
References
- Wizenty J and Sigal M. Helicobacter pylori, microbiota and gastric cancer – principles of microorganism-driven carcinogenesis. Nat Rev Gastroenterol Hepatol. 2025; 22(5):296-313.
- Fischer AS, Müllerke S, Arnold A, Heuberger J, Berger H, Lin M, […], Horst D, Tacke F, Sigal M. R-spondin/YAP axis promotes gastric oxyntic gland regeneration and Helicobacter pylori-associated metaplasia in mice. J Clin Invest. 2022; 132(21):e151363.
- Wizenty J, Müllerke S, Kolesnichenko M, Heuberger J, Lin M, Fischer AS, […], Berger H, Tacke F, Sigal M. Gastric stem cells promote inflammation and gland remodeling in response to Helicobacter pylori via Rspo3-Lgr4 axis. EMBO J. 2022; 41(13):e109996.
- Kapalczynska M, Lin M, Maertzdorf J, Heuberger J, Muellerke S, Zuo X, […], Tacke F, Meyer TF, Sigal M. BMP feed-forward loop promotes terminal differentiation in gastric glands and is interrupted by H. pylori-driven inflammation. Nat Commun. 2022; 13(1):1577.
Polyamine metabolism as a cornerstone of immune health during aging using nutritional interventions
Principal Investigator

Dr. Sebastian Hofer
Scientific interest within the context of the graduate college:
Healthy aging is increasingly recognized as a dynamically regulated state requiring active molecular maintenance in a concert of organ systems, among which the immune system takes a conducting role. The Else Kröner-Promotionskolleg “Re-Thinking Health” highlights the importance of studying resilience mechanisms that allow the body to adapt to environmental stressors to maintain (immune) homeostasis. Recent work from the PI lab (Alsaleh et al. 2020; 2025; Zhang et al. 2019; Puleston et al. 2014) and the Co-PI (Hofer et al. 2024) has revealed that the polyamine metabolism, traditionally viewed in the context of cellular proliferation, plays a key role in autophagy regulation and immune cell function. Importantly, we have shown that polyamine metabolism is a critical weak point in maintaining autophagy in advanced age (Hofer et al. 2022). Our recent discoveries have demonstrated that feeding-fasting rhythms in humans increase systemic and cellular polyamine levels and that this is crucial for regulating autophagy (Hofer et al. 2024). A central feature of this pathway is the hypusination of the eukaryotic initiation factor 5A (eIF5A), a unique post-translational modification in which the polyamine spermidine is covalently linked to a specific lysine residue, allowing cells to access a special subset of proteins that are crucial for autophagy, mitochondrial function, and immune cell fate. However, the extent to which the nutrition-polyamine axis regulates immune cell functionality in response to exogenous stimuli (e.g., vaccines, infections) has not been investigated. Therefore, this project aims to systematically dissect the role of the polyamine–hypusination axis in human immune cells under nutritional modulation, with a strong focus on clinical relevance and translational applicability.
Project description:
Aging is associated with an increase in chronic inflammation and a decline in immune function, often termed immunosenescence, which leads, inter alia, to poor vaccine responses and higher infection rates. One central metabolic axis involved in immune cell activation and differentiation is polyamine metabolism. Polyamines (putrescine, spermidine, and spermine) support cellular function via translation regulation, chromatin remodeling, and autophagy regulation. While polyamines have shown promise to counteract manifold aspects of aging in murine models, their role in human immune homeostasis, particularly under varying nutritional conditions, remains poorly defined. Previously, the Simon lab has reported that declining polyamine levels lead to autophagy deficiencies, which are partially causal for diminished T and B cell functionality and vaccine responses (Zhang et al. 2019; Alsaleh et al. 2020). Based on this, clinical trials were conceptualized to counteract the aging-polyamine axis via dietary spermidine supplementation (Alsaleh et al. 2025). Further, recent data suggests that short-term fasting activates polyamine synthesis in cells and humans (Hofer et al. 2024) and could thus be employed as an acute intervention to ramp up polyamine levels before, e.g., a vaccine challenge. How this plays out at the granular level of human immune cells has not been addressed thus far. Preliminary data in the Simon lab suggest that acute fasting modulates the downstream effector of polyamines, activated (thus hypusinated) eIF5A, across different immune cell types. This could be one mechanism by which fasting supports autophagy in the human immune system and potentially modulates the immune system’s ability to react to vaccines and infections. Based on this, we will carry out a clinical trial to test the hypothesis that four weeks of fasting could prime the aging immune system to enhance vaccine outcomes. Therefore, this project aims to characterize polyamine metabolism in human immune cells using clinical samples from ongoing dietary intervention trials, with a focus on aging, fasting, and vaccine responses.
Aim 1: We will systematically profile polyamine levels and hypusinated eIF5A across major immune cell subsets (T and B cells, monocytes, and innate lymphoid cells) using a recently established flow cytometry-based assay for hypusinated eIF5A. This will be accompanied by testing polyamine uptake, gene expression analysis, and intracellular metabolite levels, thus giving a holistic overview of the metabolic pathway. This analysis will be performed on fresh PBMCs from healthy donors of different ages to establish a reference atlas of polyamine metabolism across immune populations and to detect baseline inter-individual variability relevant to aging and nutritional state.
Aim 2: Here, we will explore how different immune cell types respond to nutritional cues, focusing on short-term fasting (in humans) and amino acid restriction (in vitro). Using in vitro culture systems and PBMCs from fasted healthy volunteers (collaboration in place), we will assess the dynamics of the polyamine metabolism and hypusination. This will reveal nutrient-responsive immune cell types and provide mechanistic insight into how dietary inputs translate into metabolic changes in the immune system. We will correlate these data with autophagic flux assays and physiological parameters of the volunteers.
Aim 3: We will analyze selected immune cell types from a randomized controlled trial of early Time-Restricted Eating (TRE) followed by influenza vaccination in older individuals (>60 years: a threshold after which vaccine responsiveness significantly drops). Using flow cytometry, metabolomics, and single-cell RNA-seq, we will assess whether the nutritional intervention alters polyamine metabolism and eIF5A hypusination in these target populations, and how these changes correlate with vaccine response and functional immune metrics (e.g., cytokine production, memory formation).
Aim 4: Finally, we will assess the functional consequences of polyamine metabolism and eIF5A hypusination on immune cell performance. Using pharmacologic and genetic tools to block hypusination or polyamine biosynthesis, we will evaluate how these pathways affect immune cell activation, autophagy, and effector function in the context of altered nutrient availability. These mechanistic experiments will be performed in vitro using primary human immune cells.
Together, this project will identify which immune cells depend on polyamine-driven mechanisms for maintaining function during nutritional stress, how these mechanisms change with age, and whether they can be modulated by clinically feasible interventions like TRE.
Methodological scope. This work draws from primary human samples collected in a clinical fasting trial (to be completed early 2026), complemented by mechanistic studies in cell lines and other primary clinical samples. The candidate will apply state-of-the-art molecular biology and immunology techniques, including spectral flow cytometry, metabolomics, single-cell RNA-seq, and autophagy assays. Mechanistic validation will be pursued through in vitro immune assays. Approximately 90% of the work will focus on human translational research.
Outlook. The successful clinician-scientist candidate in this project will generate clinically relevant insights into how polyamine metabolism supports immune resilience and is influenced by varying nutritional conditions. This work will contribute to an emerging paradigm shift in immunology, moving away from merely reacting to diseases toward health preservation through lifestyle-adapted, yet molecularly well-characterized strategies. The knowledge generated will also enhance our understanding of the significant inter-individual differences in systemic and intracellular polyamine metabolism, which could potentially serve as predictive indicators for immune health across aging trajectories.
References
- Alsaleh G, Ali M, Kayvanjoo A, Liu F, Bibi S, Luo L, […], Klenerman P, Jones L, Simon AK. Spermidine Mitigates Immune Cell Senescence, Enhances Autophagy, and Boosts Vaccine Responses in Healthy Older Adults. Research Square (PrePrint). 2025. https://doi.org/10.21203/rs.3.rs-5686388/v1.
- Alsaleh G, Panse I, Swadling L, Zhang H, Richter FC, Meyer A, […], Klenerman P, Green C, Simon AK. Autophagy in T Cells from Aged Donors Is Maintained by Spermidine and Correlates with Function and Vaccine Responses. eLife. 2020; 9:e57950.
- Hofer SJ, Daskalaki I, Bergmann M, Friščić J, Zimmermann A, Mueller MI, […], Tavernarakis N, Kroemer G, Madeo F. Spermidine Is Essential for Fasting-Mediated Autophagy and Longevity. Nat Cell Biol. 2024; 26(9):1571-1584.
- Hofer SJ, Simon AK, Bergmann M, Eisenberg T, Kroemer G, Madeo F. Mechanisms of Spermidine-Induced Autophagy and Geroprotection. Nat Aging. 2022; 2(12):1112-1129.
- Puleston DJ, Zhang H, Powell TJ, Lipina E, Sims S, Panse I, […], Townsend AR, Klenerman P, Simon AK. Autophagy Is a Critical Regulator of Memory CD8+ T Cell Formation. eLife. 2014; 3:e03706.
- Zhang H, Alsaleh G, Feltham J, Sun Y, Napolitano G, Riffelmacher T, […], Balabanov S, Mellor J, Simon AK. Polyamines Control eIF5A Hypusination, TFEB Translation, and Autophagy to Reverse B Cell Senescence. Mol Cell. 2019; 76(1):110-125.e9.
Disease phenotypes in Crohn’s disease – Does the key lie in the mucosal microenvironment?
Principal Investigator

Dr. Lea-Maxie Haag
Dr. Malte Lehmann
Scientific interest within the context of the graduate college:
Based at the Department of Gastroenterology, Infectious Diseases and Rheumatology at Campus Benjamin Franklin, Charité – Universitätsmedizin Berlin, our main clinical and research focus are inflammatory bowel diseases (IBD). Crohn’s disease (CD), one of the primary forms of IBD, exhibits various clinical phenotypes, including inflammatory (B1), stricturing (B2), and fistulizing (B3) disease phenotypes. We aim at understanding the biological differences between these phenotypes at the level of the mucosal microenvironment. With our large endoscopy unit and outpatient clinic, as well as gastroenterology ward, we have access to biological samples like intestinal biopsies along with the corresponding clinical data.
Project description:
What is the underlying mechanism and biology leading to the different disease phenotypes in Crohn’s disease?
Within this project, we aim to better understand the mechanisms responsible for the development of the different disease phenotypes in CD. We aim to analyze the intestinal microenvironment on a single-cell level by metabolic imaging mass cytometry. This enables us to investigate activity and metabolic states of intestinal structural as well as innate and adaptive immune cells. Moreover, we will focus on the relevance of cell-cell interactions and cellular neighborhoods, which define but are also common to distinct disease phenotypes (Figure 1). The proposed project will be associated with the clinical study ‘InFlame’ (DRKS00031203), which is already running since early 2023. All protocols and methods are established and running in the lab and have been approved by the ethics board of the Charité – Universitätsmedizin Berlin. The existing study team and the ongoing recruitment process guarantee intensive support and the provision of all necessary infrastructure.
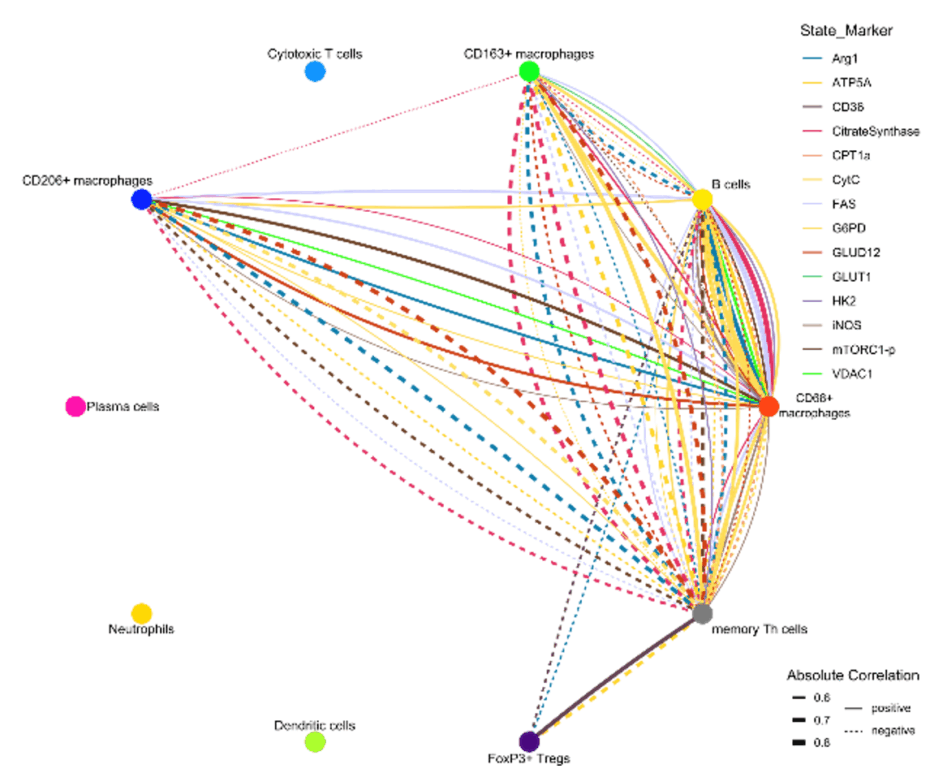
Figure 1. Iconography of correlations of state marker expression to the distance between immune cell clusters. Only correlations with r > 0.5 or < 0.5 and p < 0.05 are shown. The color of the lines indicate the respective state marker, the width of the line the absolute value of correlation. Straight lines indicate positive correlations, doted lines negative correlations.
WP1. Crohn’s Disease Cohort Recruitment. Completing the B1-B2-B3 Crohn’s disease cohort (recruitment already ongoing).
WP2. Imaging Mass Cytometry of Intestinal Tissue to Study Crohn’s Disease. Performing imaging mass cytometry on intestinal tissue (using Formalin fixed paraffin embedded biopsies) and analyses of the different disease phenotypes in Crohn’s disease.
References
- Rieder F, Zimmermann EM, Remzi FH, Sandborn WJ. Crohn’s disease complicated by strictures: a systematic review. Gut. 2013; 62(7):1072-1084.
- Rieder F, Mukherjee PK, Massey WJ, Wang Y, Fiocchi C. Fibrosis in IBD: from pathogenesis to therapeutic targets. Gut. 2024; 73(5):854-866.
- Lehmann M, Weixler B, Elezkurtaj S, Loddenkemper C; TRR241 IBDome Consortium; Kühl AA, Siegmund B. Spatial Single Cell Profiling Using Imaging Mass Cytometry: Inflammatory Versus Penetrating Crohn’s Disease. J Crohns Colitis. 2024; 18(8):1305-1318.
- Lehmann M, Allers K, Heldt C, Meinhardt J, Schmidt F, Rodriguez-Sillke Y, […], Weidinger C, Kühl AA, Siegmund B. Human small intestinal infection by SARS-CoV-2 is characterized by a mucosal infiltration with activated CD8+ T cells. Mucosal Immunol. 2021; 14(6):1381-1392.
- Avery EG*, Haag LM*, McParland V, Kedziora SM, Zigra GJ, Valdes DS, […], Siegmund B, Wiig H, Müller DN. Intestinal interstitial fluid isolation provides novel insight into the human host-microbiome interface. Cardiovasc Res. 2025; 121(5):803-816. *equal contribution
- Stankey CT*, Bourges C*, Haag LM*, Turner-Stokes T, Piedade AP, Palmer-Jones C, […], Wallace C, Thomas DC, Lee JC. A disease-associated gene desert directs macrophage inflammation through ETS2. Nature. 2024; 630(8016):447-456. *equal contribution
Identification of a gut microbiota signature in patients with drug-resistant epilepsy upon ketogenic diet treatment
Principal Investigator

Dr. Laura Díaz Marugán
Scientific interest within the context of the graduate college:
This project aims to uncover the intricate relationship between diet, gut microbes, and drug-resistant epilepsy (DRE). Focusing on the ketogenic diet (KD), the research seeks to identify specific gut bacteria associated with improved brain health in KD responders. The objectives include investigating the bacterial signature in a large cohort of KD responder and non-responder people, isolating potentially beneficial bacteria, and studying their metabolic functions. The study aims to understand how the gut microbiota influences the success of KD treatment, potentially serving as a biomarker for treatment efficacy. By bridging the gap between diet, gut microbes, and neurological conditions, this research may pave the way for personalized interventions in managing drug-resistant epilepsy.
This project stands out for its strong clinical and experimental approach in close collaboration with Prof. Dr. Angela Kaindl from the Clinic for Pediatrics and Neurology, Charité. The successful candidate will benefit from a training in which he/she will be able to put into practice his/her knowledge of the medical field and learn concepts, techniques and strategies of experimental research in the laboratory, paving the way to better understand how we could modulate the course of epilepsy via microbiota-diet modulation.
In this research project, the successful candidate will be able to:
- Manage human sampling collection for research purposes.
- Learn about the ketogenic diet as a therapy in the clinics.
- Learn about patient’s group stratification according to the different profiles in response to the treatment.
- Prepare and analyse questionnaires to track dietary habits, life-style habits and quality of life.
- Extract and quantify genomic DNA from human stools.
- Culture bacteria from human stools and characterize them via full genome sequence.
- Get knowledge in microbial composition bioinformatic analysis.
- Process serum samples for proteomic analysis.
Project description:
Epidemiology and social impact of epilepsy and drug-resistant epilepsy (DRE). Epilepsy is one of the most common neurological disorders, affecting more than 50 million people worldwide and 6 million people in Europe (Singh & Sander, 2020; Sirven, 2015). It affects people of all ages and is characterized by epileptic seizures (Berg et al., 1999; Fiest et al., 2017; Guerrini, 2006; Sirven, 2015). Uncontrolled seizures can lead to developmental delay, cognitive deficits, memory and learning difficulties. They can also cause sudden unexpected death in people with epilepsy (SUDEP) (Sperling, 2004). Current treatments include anti-seizure medicines (ASM), diet, epilepsy surgery and stimulation devices. ASM can control seizures in about two-thirds of people with epilepsy. However, one-third of patients with epilepsy cannot be controlled by two or more correctly selected and dosed ASM, what is called drug-resistant epilepsy (DRE) (Kwan et al., 2009, 2011; Picot et al., 2008; Sultana et al., 2021).
The microbiota-gut-brain axis: a modulator for epileptogenesis. The causes of epilepsy include structural, genetic, infectious, metabolic and immune factors (Guerri et al., 2020; Matin et al., 2015; Vezzani et al., 2016; Oliver et al., 2023; Rho & Boison, 2022). The gut-brain axis (GBA) has also been implicated in this respect (Iannone et al., 2019). Gut microbiota produces a variety of compounds that can affect brain function, such as neurotransmitters and metabolites, like short-chain fatty acids (SCFAs) (Chen et al., 2021; Dalile et al., 2019; Silva et al., 2020), some of which have shown anticonvulsant effects in rodents (De Caro et al., 2019; Mu et al., 2022; Olson et al., 2018). In patients, studies have confirmed that bacterial-based approaches, such as probiotics or microbiota-modifying treatments, can reduce seizure frequency. Indeed, the pilot usage of a mixture of probiotics (eight bacterial subspecies of Lactobacillus, Bacteroides and Streptococcus) reduced seizure frequency and improved the quality of life in patients with DRE (Gómez-Eguílaz et al., 2018). Similarly, fecal microbiota transplantation (FMT) for the treatment of Crohn’s disease (CD) symptoms ameliorated seizures frequency in a 22-year-old woman with a 17-year history of epilepsy, who stopped taking ASM after the FMT (He et al., 2017). Despite these promising results, the mechanisms by which the microbiota may affect the development of seizures in patients are completely unclear.
Ketogenic diet in the management of DRE. Alternative treatments for DRE include dietary treatments such as the ketogenic diet (KD) (Hemingway et al., 2001; Lyons et al., 2020). KD is a diet based on a high fat intake accompanied by moderate protein, and very low or no carbohydrate content, usually in a 3:1 or 4:1 ratio of caloric intake from lipids to both protein and carbohydrate together, and with a normal total energy intake. Restricting carbohydrate intake forces the metabolism to replace the preferred energy source, glucose, with fatty acids, which are oxidized in the liver to form ketone bodies. Although the classical KD has been used in epilepsy for many years and its efficacy in reducing seizures has been confirmed in several studies, especially when introduced in young age (Martin-McGill et al., 2020), the mechanism behind its action is not well understood in humans. In particular, it is not known why this dietary intervention has an anti-seizure effect in a proportion of the patients, while in others there is poor or no effect (Martin-McGill et al., 2020) or even worsening of seizure frequency or appearance of adverse effects (Cai et al., 2017; Newmaster et al., 2022; Yan et al., 2018). Although the exact mechanism is not yet known, several research studies in both mice and human have shown a link between the gut microbiota and the effects of the KD, topic that we recently discussed (Díaz-Marugan et al., 2024).
Hypothesis. Changes in the gut microbial composition by dietary intervention have a significant impact on the CNS function during the development of pathological conditions, such as seizures, via metabolites or immune mediators.
Therefore, this proposal aims to provide unprecedented insights into the molecular characteristics of commensal microbes after ketogenic diet that determine seizure improvement or worsening in different patients.
To achieve the overall aim, I have defined two objectives:
- Determine the (i) faecal microbiota composition and (ii) stool and blood metabolomic profile of children with DRE and treated with KD or with a non-KD diet compared to healthy children.
- Isolate and characterise the metabolic functions of commensal bacteria that thrive in children with DRE and treated with KD and show seizure improvement, no effect on seizures or worsening after KD introduction.
The successful candidate for this fellowship will focus on addressing Aim 1 and 2.
Aim 1: Human KD-associated microbiota and metabolomics. Hypothesis: In humans, the KD is associated with an overgrowth of gut bacteria capable of secreting neuroprotective metabolites with an anti-seizure activity. Aim: To investigate the (i) faecal microbiota composition and (ii) stool and blood metabolomic profile of children with drug-resistant epilepsy and treated with KD or with a non-KD diet, compared to healthy children. Work plan (WP): To address this first aim, we will recruit the following participants: (i) with drug-resistant epilepsy (DRE) for which KD is recommended. They will start the treatment with KD within an inpatient study (KD) (group 1), (ii) with DRE and non-KD diet (group 2), (iii) healthy children on a non-KD diet (group 3).
From group 1, several faecal samples, blood samples and questionnaire responses are collected. From group 2, a faecal sample, a blood sample and answers to a questionnaire are collected. A faecal sample and questionnaire responses will be collected from group 3. There will be 50 participants recruited in each group (within each group there will be a 1:1 male to female ratio).
The following analyses are planned:
Analysis 1. Validated food frequency and lifestyle questionnaires for each study participant. Dietary information must be recorded for the last five days prior to study recruitment. Dietary and lifestyle information will be recorded by the study dietitian in an epilepsy database developed by the Charité and will be included in our analysis.
Analysis 2. Clinical characterisation of epilepsy for the first two groups. Data collected during routine epilepsy care will be recorded in a standardised questionnaire: manifestation, seizures, seizure calendar, epilepsy / epilepsy syndrome, possible structural, genetic and autoimmune causes, developmental diagnostics. All patients are followed clinically by Prof. Kaindl’s medical team and all patient data are recorded in an epilepsy database developed by the Charité. Other important information about the patients’ treatment with anticonvulsants drugs is also recorded and taken into account in our analysis.
Analysis 3. Stool microbiome composition and metabolomics for all groups. Stool samples will be collected cross-sectionally from each group and longitudinally from group 1. Stool samples will be collected from individuals in group 1 before KD initiation, twice in the first week after KD initiation, then once two weeks after KD initiation, and then every month after KD initiation for up to six months. Faeces are freshly collected in the OMNIgene®-GUT kit, in which microbial DNA can be stored at room temperature for up to 3 months from the time of collection, prior to analysis of microbial composition using metagenomics to identify changes in microbiota composition. Fecal DNA will be prepared using the QIAamp PowerFecal Pro DNA Kit (QIAGEN) according to the manufacturer’s instructions. Sequencing will be performed on the Novaseq Xplus platform (Illumina) using the NovaSeq X Series 25B Reagent Kit (300 Cycle) (Illumina) at an average depth of 7.5 Gbases per sample. A second faecal aliquot sample will be collected in OMNImet®-GUT optimised to perform untargeted and targeted metabolomics. Amino acids, lipids, fatty acids, ketone bodies (beta-hydroxybutyrate, BHB and acetoacetate), indoles and bile acids will be quantified.
Analysis 4. Determination of serum metabolites and inflammatory parameters. Blood samples will only be collected from participants in the fist two groups for untargeted and targeted metabolomics, measurements of inflammatory mediators such as cytokines and chemokines will be performed on blood serum using the Meso Scale Discovery platform.
Aim 2. Isolation and characterisation of KD-associated human bacterial consortia. Hypothesis: The success of KD treatment in people with drug-resistant epilepsy is related to the composition and function of the gut microbiota. Aim: To isolate and characterise the metabolic functions of commensal bacteria thriving in paediatric patients with epilepsy treated with KD and showing (a) seizure improvement, (b) no seizure effect or (c) seizure worsening after KD introduction. Work plan: Ex vivo isolation and sequencing of KD-associated human bacteria. After one to 12 weeks on the new diet, some participants in group 1 will respond to the KD intervention and show an improvement in seizures (KD responders, >50% of the treated patients, with at least 50% of seizures improving -group 1A-), some will show neither improvement nor worsening of seizure evolution (KD non-responders, about 30% of the patients -group 1B-), while others will experience worsening of the disease after KD introduction(KD worse-responders, 5-10% of the patients -group 1C-). 3-5 subjects/group will be selected according to the development of seizures after KD consumption and a faecal sample will be collected within three months (for groups 1B and 1C) or six months (for group 1A) after the start of KD. This sample will be collected in the GutAlive (MicroViable Therapeutics) kit and sent immediately to the laboratory. This aliquot will be used for in vitro culture of the bacteria. in an anaerobic chamber. Each bacterial culture will be tested by Gram staining to assess the purity, and the genomic DNA is extracted to perform whole genome sequencing on the Pacific Biosciences (PacBio) Sequel machine at the Genomics Core Facility (MDC, Berlin). In this way, we will have the complete genome sequence of the identified bacteria, which will allow us to know all the genes expressed by each bacterium. This approach will allow us to select the bacteria that can be grown in vitro from the different groups of participant.
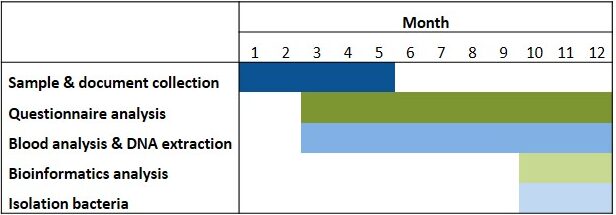
Table 1. Timeline. Isolation of bacteria will be possible depending on the development of the aim 1 of the research project.
Abbreviations. ASM: anti-seizure medicines; DRE: drug-resistant epilepsy; FatsQ: fats-type questionnaire; FFQ: food frequency questionnaire; KD: ketogenic diet; LifeSQ: lifestyle questionnaire; QOLIE 31: quality of life in epilepsy questionnaire
References
- Afrizal A, Hitch TCA, Viehof A, Treichel N, Riedel T, Abt B, […], Kohlheyer D, Overmann J, Clavel T. Anaerobic single-cell dispensing facilitates the cultivation of human gut bacteria. Environ Microbiol. 2022; 24(9):3861-3881.
- Berg AT, Shinnar S, Levy SR, Testa FM. Newly diagnosed epilepsy in children: presentation at diagnosis. Epilepsia. 1990; 40(4):445-452.
- Cai QY, Zhou ZJ, Luo R, Gan J, Li SP, Mu DZ, Wan CM. Safety and tolerability of the ketogenic diet used for the treatment of refractory childhood epilepsy: a systematic review of published prospective studies. World J Pediatr. 2017; 13(6):528-536.
- Chen Y, Xu J, Chen Y. Regulation of neurotransmitters by the gut microbiota and effects on cognition in neurological disorders. Nutrients. 2021; 13(6):1-21.
- Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat Rev Gastroenterol Hepatol. 2019; 16(8):461-478.
- De Caro C, Leo A, Nesci V, Ghelardini C, di Cesare Mannelli L, Striano P, […], Citraro R, De Sarro G, Russo E. Intestinal inflammation increases convulsant activity and reduces antiepileptic drug efficacy in a mouse model of epilepsy. Sci Rep. 2019; 9(1):1-10.
- Díaz-Marugán L, Rutsch A, Kaindl AM, Ronchi F. The impact of microbiota and ketogenic diet interventions in the management of drug-resistant epilepsy. Acta Physiol (Oxf). 2024; 240(3):e14104.
- Fiest KM, Sauro KM, Wiebe S, Patten SB, Kwon CS, Dykeman J, Pringsheim T, Lorenzetti DL, Jetté N. Prevalence and incidence of epilepsy. Neurology. 2017; 88(3):296-303.
- Guerri G, Castori M, D’Agruma L, Petracca A, Kurti D, Bertelli M. Genetic analysis of genes associated with epilepsy. Acta Biomed. 2020; 91(13-S):e2020005.
- Guerrini R. Epilepsy in children. Lancet. 2006; 367(9509):499-524.
- Gómez-Eguílaz M, Ramón-Trapero JL, Pérez-Martínez L, Blanco JR. The beneficial effect of probiotics as a supplementary treatment in drug-resistant epilepsy: A pilot study. Benef Microbes. 2018; 9(6):875-881.
- He Z, Cui BT, Zhang T, Li P, Long CY, Ji GZ, Zhang FM. Fecal microbiota transplantation cured epilepsy in a case with Crohn’s Disease: The first report. World J Gastroenterol. 2017; 23(19):3565-3568.
- Hemingway C, Freeman JM, Pillas DJ, Pyzik PL. The ketogenic diet: A 3- to 6-year follow-up of 150 children enrolled prospectively. Pediatrics. 2001; 108(4):898-905.
- Iannone LF, Preda A, Blottière HM, Clarke G, Albani D, Belcastro V, […], Zara F, Russo E, Striano P. Microbiota-gut brain axis involvement in neuropsychiatric disorders. Expert Rev Neurother. 2019; 19(10):1037-1050.
- Kwan P, Arzimanoglou A, Berg AT, Brodie MJ, Allen Hauser W, Mathern G, […], Perucca E, Wiebe S, French J. Definition of drug resistant epilepsy: Consensus proposal by the ad hoc Task Force of the ILAE Commission on Therapeutic Strategies. Epilepsia. 2009; 51(6):1069-1077.
- Kwan P, Schachter SC, Brodie MJ. Drug-resistant epilepsy. N Engl J Med. 2011; 365(10):919-926.
- Lyons L, Schoeler NE, Langan D, Cross JH. Use of ketogenic diet therapy in infants with epilepsy: A systematic review and meta-analysis. Epilepsia. 2020; 61(6):1261-1281.
- Matin N, Tabatabaie O, Falsaperla R, Lubrano R, Pavone P, Mahmood F, […], Di Mauro P, Cocuzza S, Vitaliti G. Epilepsy and innate immune system: a possible immunogenic predisposition and related therapeutic implications. Hum Vaccin Immunother. 2015; 11(8):2021-2029.
- Martin-McGill KJ, Bresnahan R, Levy RG, Cooper PN. Ketogenic diets for drug-resistant epilepsy. Cochrane Database Syst Rev. 2020; 6(6):CD001903.
- Mu C, Nikpoor N, Tompkins TA, Choudhary A, Wang M, Marks WN, Rho JM, Scantlebury MH, Shearer J. Targeted gut microbiota manipulation attenuates seizures in a model of infantile spasms syndrome. JCI Insight. 2022; 7(12):e158521.
- Newmaster K, Zhu Z, Bolt E, Chang RJ, Day C, Mhanna A, […], Mainali G, Carney PR, Naik S. A Review of the Multi-Systemic Complications of a Ketogenic Diet in Children and Infants with Epilepsy. Children. 2022; 9(9):1372.
- Oliver KL, Scheffer IE, Bennett MF, Grinton BE, Bahlo M, Berkovic SF. Genes4Epilepsy: an epilepsy gene resource. Epilepsia. 2023; 64(5):1368-1375.
- Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY. The Gut Microbiota Mediates the Anti-Seizure Effects of the Ketogenic Diet. Cell. 2018; 173(7):1728-1741.e13.
- Picot MC, Baldy-Moulinier M, Daurès JP, Dujols P, Crespel A. The prevalence of epilepsy and pharmacoresistant epilepsy in adults: A population-based study in a Western European country. Epilepsia. 2008; 49(7):1230-1238.
- Rho JM and Boison D. The metabolic basis of epilepsy. Nat Rev Neurol. 2022; 18(6):333-347.
- Silva YP, Bernardi A, Frozza RL. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front Endocrinol (Lausanne). 2020; 11:25.
- Singh G and Sander JW. The global burden of epilepsy report: Implications for low- and middle-income countries. Epilepsy Behav. 2020; 105:106949.
- Sirven JI. Epilepsy: A Spectrum Disorder. Cold Spring Harb Perspect Med. 2015; 5(9):a022848.
- Sperling MR. The consequences of uncontrolled epilepsy. CNS Spectr. 2004; 9(2):98-101,106-109.
- Sultana B, Panzini MA, Veilleux Carpentier A, Comtois J, Rioux B, Gore G, […], Jetté N, Josephson CB, Keezer MR. Incidence and Prevalence of Drug-Resistant Epilepsy: A Systematic Review and Meta-analysis. Neurology. 2021; 96(17):805-817.
- Vezzani A, Fujinami RS, White HS, Preux PM, Blümcke I, Sander JW, Löscher W. Infections, inflammation and epilepsy. Acta Neuropathol. 2016; 131(2):211-234.
- Yan N, Xin-Hua W, Lin-Mei Z, Yi-Ming C, Wen-Hui L, Yuan-Feng Z, Shui-Zhen Z. Prospective study of the efficacy of a ketogenic diet in 20 patients with Dravet syndrome. Seizure. 2018; 60:144-148.
Dissecting the functional impact of somatic mutations on NK cell memory
Principal Investigator

Dr. Timo Rückert
Scientific interest within the context of the graduate college:
Within the framework of the graduate college “Re-Thinking Health,” our goal is to deepen our understanding of how the innate immune system contributes to both the maintenance of health and the development of chronic inflammatory diseases. While persistent immune memory is essential for long-term protection against infections, it can also hinder therapeutic success in chronic inflammatory conditions. Traditionally associated with the adaptive immune system, immune memory has recently been observed in innate lymphoid cells (ILCs), particularly Natural killer (NK) cells. However, the mechanisms underlying the establishment and maintenance of NK cell memory, as well as its functional consequences for health and disease, remain only partially understood. By investigating ILC and NK cell responses to viral infections and chronic inflammatory environments, we aim to uncover the extrinsic signals, intrinsic cellular characteristics, and molecular networks that drive memory formation and persistence. This integrative approach will not only expand our understanding of innate immune memory but may also reveal actionable targets to counteract pathological memory or harness memory persistence for improved cell-based therapies.
Project description:
Natural killer (NK) cells are innate lymphocytes that play critical roles in immune defense against viral infections and cancer. Although classically regulated by germline-encoded receptors, NK cells can undergo clonal expansion and epigenetic remodeling, acquiring memory-like properties that enhance their responsiveness to specific stimuli – particularly following infection with cytomegalovirus (CMV). Our recent findings reveal the long-term persistence of highly expanded CMV-specific NK cell clones carrying somatic mutations. These mutations are enriched in cancer driver genes and immune-related pathways, suggesting functional consequences that may contribute to clonal selection and persistence. This challenges the conventional view of NK cell regulation and introduces somatic genetic variability as a novel mechanism influencing immune cell fitness. The goal of this project is to investigate the molecular and functional consequences of selected somatic mutations on NK cell proliferation, survival, and effector functions. Through a combination of genetic perturbation and cellular assays, we aim to elucidate how these variants contribute to NK cell clonal expansion and long-term memory, and how they may enhance antiviral responses. These studies will advance our understanding of NK cell biology and could inform novel strategies to optimize NK cell-based immunotherapies for infection and cancer.
Aim 1: Optimization and validation of targeted gene editing. Building on up-and-running gene editing methods (CRISPR/Cas9, Cytosine-base editing), we will optimize and validate efficient gene-targeting approaches in primary NK cells to model selected loss-of-function candidates.
Aim 2: Characterizing the functional impact of candidate mutations. We will utilize established in vitro assays to assess the consequences of candidate variants on NK cell effector functions, proliferation, and survival in response to different stimuli. If time allows, these assays might be extended by a murine model of CMV infection.
References
- Rückert T, Lareau CA, Mashreghi MF, Ludwig LS, Romagnani C. Clonal expansion and epigenetic inheritance of long-lasting NK cell memory. Nat Immunol. 2022; 23(11):1551-1563.
- Rückert T and Romagnani C. Extrinsic and intrinsic drivers of natural killer cell clonality. Immunol Rev. 2024; 323(1):80-106.
- Hammer Q, Rückert T, Borst EM, Dunst J, Haubner A, Durek P, […], Mashreghi MF, Messerle M, Romagnani C. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat Immunol. 2018; 19(5):453-463.
Fibrosis in Crohn’s disease – what are the drivers in the mucosal microenvironment?
Prinicipal Investigator

Dr. Lea Maxie Haag
Scientific interest within the context of the graduate college:
Based at the Department of Gastroenterology, Infectious Diseases and Rheumatology at Campus Benjamin Franklin, Charité – Universitätsmedizin Berlin and at the Max Delbrück Center (MDC) in Berlin, our main clinical and research focus are chronic inflammatory diseases like Inflammatory bowel diseases (IBD). Crohn’s disease (CD), one of the main forms of IBD, has different clinical phenotypes, including an inflammatory (B1) as well as a stricturing (B2) disease phenotype, in which patients develop fibrotic stenoses of the intestine. Developing a molecular understanding of why some patients suffer from the formation of fibrotic strictures, while others do not, is still not understood. By investigating the mucosal microenvironment, we aim to establish a holistic understanding of the process of fibrotic stricture formation in CD to find answers to the driving question: ‘Stricturing in Crohn’s disease – what are the drivers in the mucosal microenvironment?’
Project description:
What is the underlying mechanism leading to the formation of fibrotic strictures in Crohn’s disease? Why do some patients develop a stricturing disease phenotype while others do not?
Within this project, we aim to better understand mechanisms responsible for the development of a stricturing phenotype. We hypothesize that the mucosal microenvironment in patients with an inflammatory CD phenotype (B1) differs from the microenvironment found in patients with a stricturing phenotype (B2). Therefore, the mandatory prerequisite is to deeply characterize the different disease phenotypes. The proposed project will be associated to the clinical study ‘InFlame’ (DRKS00031203), which is already running since early 2023. We have developed a ‘mucosa phenotyping pipeline’ (Figure 1) with high-throughput methods to analyze the intestinal microenvironment e.g., interstitial fluid and mucosal microbiota from biopsies, 3D histology and imaging mass cytometry as well as components of the peripheral immune system via blood samples, to comprehensively phenotype IBD patients and healthy individuals.
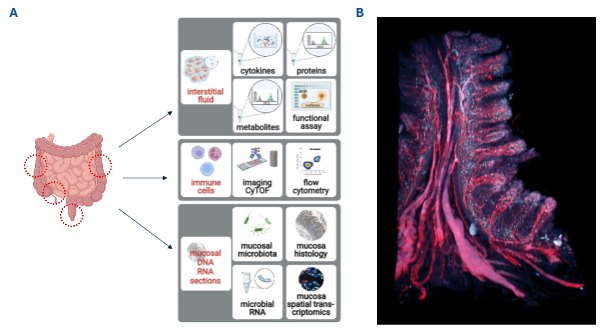
Figure 1. Mucosa Phenotyping pipeline: A. Biopsies will be obtained during endoscopy at the indicated red circles and handled as described in detail in the main text. B. Human intestinal biopsy stained in red with nanobodies against CD31 (vessels) and podoplanin (lymphatics) and blue against CD8 T cells.
All methods are established and running in the lab. The existing study team and the ongoing recruitment process guarantee intensive support and the provision of all necessary infrastructure.
WP1. Identification of Crohn’s Disease Inflammatory Subtypes. Identify Crohn’s disease patients already recruited who either have an inflammatory B1-phenotype or a stricturing B2-phenotype.
WP2. Deep phenotyping by our mucosa phenotyping platform. Biopsies will be used for (i) isolation of IF, (ii) phenotyping of the immune- cells including 3D histology (iii) analysis of mucosal microbiota. This approach will result in an in-depth spatiofunctional and spatiomechanistic atlas for microbes, metabolites, proteins, cytokines, immune cells, and transcriptomes from different segments of the intestinal tract.
References
- Avery EG*, Haag LM*, McParland V, Kedziora SM, Zigra GJ, Valdes DS, […], Siegmund B, Wiig H, Müller DN. Intestinal interstitial fluid isolation provides novel insight into the human host-microbiome interface. Cardiovasc Res. 2025; 121(5):803-816. *equal contribution
- Stankey CT*, Bourges C*, Haag LM*, Turner-Stokes T, Piedade AP, Palmer-Jones C, […], Wallace C, Thomas DC, Lee JC. A disease-associated gene desert directs macrophage inflammation through ETS2. Nature. 2024; 630(8016):447-456. *equal contribution
- Rieder F and Zimmermann EM. Crohn’s disease complicated by strictures: a systematic review. Gut. 2013; 62(7):1072-1084.
- Rieder F and Murkherjee PK. Fibrosis in IBD: from pathogenesis to therapeutic targets. Gut. 2024; 73(5):854-866.
Impact of neutrophil serine proteases on mucus properties and function in in muco-obstructive lung diseases
Principal Investigator

Dr. Christine K. Wong
Scientific interest within the context of the graduate college:
Our airways are covered by a thin mucus layer that plays a pivotal role in maintaining lung health and homeostasis. In healthy, this mucus layer entraps constantly inhaled pathogens, pollutants and irritants that are then removed from the lungs by beating cilia on airway surfaces that facilitate mucocilary clearance, which constitutes an important innate defense mechanism of the lung. In chronic muco-obstructive lung diseases such as cystic fibrosis (CF) and bronchiectasis the viscoelastic properties of airway mucus are characteristically altered.1,2 Increased viscosity and elasticity of the mucus in these diseases leads to impaired mucociliary clearance, which in turn leads to airway mucus plugging, chronic infection with Pseudomonas aeruginosa and other bacterial pathogens, and chronic neutrophilic inflammation. Neutrophils express a group of proteases called neutrophil serine proteases (NSP), including neutrophil elastase (NE), protease 3 (PR3) and cathepsin G (CG). It is well established that increased activity of these NSPs leads to the degradation of endogenous anti-proteases, which in turn causes a protease/anti-protease imbalance that plays a central role in the pathogenesis of progressive structural lung damage in CF and bronchiectasis.3,4 Preliminary data from our group suggest that these proteases may also change the viscoelastic properties and function of the mucus by cleavage of the mucins (MUC5B and MUC5AC) that form the mucus layer. However, a systemic evaluation of the effects of NSPs on mucus properties has not been performed and whether proteolytic degradation of mucus is beneficial or detrimental in muco-obstructive lung diseases remains unknown. Finding answers to these questions is also relevant in the context of current development of novel therapies for CF and bronchiectasis that inhibits NSP activity5 and may thereby also have an effect on mucus properties and mucociliary clearance.
Project description:
The overall aim of this project is to characterize the impact of NSPs on mucus properties and function in muco-obstructive lung diseases such as in CF and bronchiectasis. Elevated levels of NSP activity are characteristically found in expectorated sputum from patients with CF and patients with bronchiectasis,1,5 and are associating with increased mucus viscoelasticity and increased proinflammatory cytokines. Three NSPs have been identified, NE, PR3 and CG that have been implicated to play a crucial role in the pathogenesis of muco-obstructive lung diseases.6,7 To study the role of the three NSPs in mucin processing and modulation of mucus properties and function in health and disease, we will perform ex vivo studies of native sputum samples from healthy people and patients with CF or bronchiectasis and in vitro studies of the native mucus layer on patient-derived airway epithelial cultures. Specifically, sputum collected from CF or bronchiectasis and healthy people will be treated with individual NSPs (NE, PR3 or CG) at various pathophysiologically relevant concentrations and the impact on mucus processing and viscoelastic properties will be determined biochemically by Western blotting and mass spectrometry and functionally by rheological measurements using a cone and plate rheometer. Since NSP activity is commonly elevated in expectorated patient sputum, these measurements will also be performed in samples that will be treated with protease inhibitors. To gain mechanistic insight, these studies in patient sputum will be complemented by studies of highly differentiated patient-derived primary airway epithelial cell cultures that allow to study the native mucus layer under near physiological condition.8 For this purpose, airway cultures will be treated with the different NSPs added at different concentrations to the mucus layer and effects on viscoelastic properties will be determined using several sophisticated imaging-based techniques such as FRAP (fluorescent recovery after photobleaching)9 and magnetic microwire rheometry (MMWR)10 that have been established in our laboratory. By stimulating primary airway cultures with proinflammatory mediators in the presence of NSPs, we can recapitulate the chronic muco-inflammatory environment characteristic of CF and bronchiectasis in vitro. These functional studies of the properties of the native mucus layer will be accompanied by biochemical studies of abundancy and processing of the airway mucins MUC5AC and MUC5B by Western blotting and mass spectrometry, as described for patient sputum samples above, and inflammatory mediators such as proinflammatory cytokines secreted by epithelial cells will be assessed by bead-based multiplexed immunoassay. Thereby, this MD thesis will apply a spectrum of molecular, cellular, and state-of-the-art imaging techniques to patient-derived sputum and airway cultures to tackle pertinent questions related to the pathogenesis and therapy of muco-obstructive lung diseases such as in CF and bronchiectasis, which have emerged as the third leading chronic lung disease worldwide with limited therapeutic options.
References
- Schaupp L, Addante A, Völler M, Fentker K, Kuppe A, Bardua M, […], Boutin S, Graeber SY, Mall MA. Longitudinal effects of elexacaftor/tezacaftor/ivacaftor on sputum viscoelastic properties, airway infection and inflammation in patients with cystic fibrosis. Eur Respir J. 2023; 62(2):2202153.
- Ramsey KA, Chen ACH, Radicioni G, Lourie R, Martin M, Broomfield A, […], Kesimer M, Boucher RC, McGuckin MA. Airway Mucus Hyperconcentration in Non-Cystic Fibrosis Bronchiectasis. Am J Respir Crit Care Med. 2020; 201(6):661-670.
- Chalmers JD, Mall MA, Chotirmall SH, O’Donnell AE, Flume PA, Hasegawa N, […], Xu JF, Shteinberg M, McShane PJ. Targeting neutrophil serine proteases in bronchiectasis. Eur Respir J. 2025; 65(1):2401050.
- Mall MA, Davies JC, Donaldson SH, Jain R, Chalmers JD, Shteinberg M. Neutrophil serine proteases in cystic fibrosis: role in disease pathogenesis and rationale as a therapeutic target. Eur Respir Rev. 2024; 33(173):240001.
- Johnson ED, Long MB, Perea L, Shih VH, Fernandez C, Teper A, […], Huang JTJ, Stobo J, Chalmers JD. Broad Immunomodulatory Effects of the Dipeptidyl-peptidase-1 Inhibitor Brensocatib in Bronchiectasis: Data from the Phase 2, Double-Blind, Placebo-controlled WILLOW Trial. Am J Respir Crit Care Med. 2025. Online ahead of print.
- Voynow JA and Shinbashi M. Neutrophil Elastase and Chronic Lung Disease. Biomolecules. 2021; 11(8):1065.
- Clancy DM, Sullivan GP, Moran HBT, Henry CM, Reeves EP, McElvaney NG, Lavelle EC, Martin SJ. Extracellular Neutrophil Proteases Are Efficient Regulators of IL-1, IL-33, and IL-36 Cytokine Activity but Poor Effectors of Microbial Killing. Cell Rep. 2018; 22(11):2937-2950.
- Balázs A, Millar-Büchner P, Mülleder M, Farztdinov V, Szyrwiel L, Addante A, […], Röhmel J, Ralser M, Mall MA. Age-Related Differences in Structure and Function of Nasal Epithelial Cultures From Healthy Children and Elderly People. Front Immunol. 2022; 13:822437.
- Balázs A, Rubil T, Wong CK, Berger J, Drescher M, Seidel K, Stahl M, Graeber SY, Mall MA. The potentiator ivacaftor is essential for pharmacological restoration of F508del-CFTR function and mucociliary clearance in cystic fibrosis. JCI Insight. 2025; 10(10):e187951.
- Braunreuther M, Liegeois M, Fahy JV, Fuller GG. Nondestructive rheological measurements of biomaterials with a magnetic microwire rheometer. J Rheol. 2023; 67(2):579-588.
Assessment of the role of pericyte loss in the pathophysiology of pulmonary arterial hypertension by organ-on-chip technology
Principal Investigator

Dr. Lasti Erfinanda
Scientific interest within the context of the graduate college:
Pulmonary arterial hypertension is a cardiopulmonary disease that is characterized by profound remodeling of the pulmonary vascular tree and has a poor prognosis if left untreated. Current therapeutic approaches rely on vasodilatory drugs and do not address the underlying cause of vascular remodeling. Investigating the pathophysiology of pulmonary arterial hypertension on a novel, in vitro microvasculature-on-chip model allows to track changes of the vascular system and the dynamics of individual cell types specifically in a temporal and spatial context that was not possible before. As such, microvasculature-on-chip models – in combination with advanced imaging modalities, state-of-the-art transcriptomic and proteomic analyses, and functional read-outs – open up unprecedented avenues for the discovery of new disease mechanisms and therapeutic targets.
Project description:
Pulmonary arterial hypertension arises from extensive remodeling of the pulmonary blood vessels, leading to elevated pulmonary vascular resistance and increased pulmonary arterial pressure. This progression places strain on the right ventricle, ultimately resulting in its dysfunction, failure, and potentially death. Pulmonary vascular remodeling in pulmonary arterial hypertension is characterized by proliferation and hypertrophy of endothelial and smooth muscle cells in pulmonary resistance vessels, and a parallel loss of pulmonary microvessels, especially in the alveolar capillary network, termed microvascular rarefaction or pruning.
Recently, this microvascular rarefaction has been linked to a loss of microvascular pericytes. Pericytes are perivascular cells that are encased within the microvascular basement membrane and assist in the maturation and stabilization of microvascular networks. To this end, pericytes communicate with endothelial cells by both direct physical contact, via gap junctions and by paracrine signalling. This interaction promotes the stabilization of endothelial cells and the maintenance of vascular barrier function. Accordingly, loss or detachment of pericytes may result in increased microvascular leak and the disintegration of microvascular networks. Recent histological analyses in lung tissue samples from PAH patients and animal models of pulmonary arterial hypertension reported an increased density of pericytes in pulmonary arterial resistance vessels, while the abundance of pericytes in the pulmonary microvasculature was markedly reduced. Based on this finding, we and others have hypothesized that in PAH, pericytes may migrate from the pulmonary capillary bed to the proximal arteries where they may integrate into the arterial media as contractile cells. Such a process would not only promote arterial remodeling and muscularization, but may also in parallel destabilize the pulmonary microvasculature causing microvasculature rarefaction. The actual effect of pericyte loss on pulmonary microvascular networks has, however, so far not been elucidated due to the lack of appropriate models and methods.
Histopathological analyses in human tissue samples or animal models are typically limited to a single timepoint and as such, cannot assess the dynamics and interactions of distinct cell types over time and space, or track the process of microvascular rarefaction over time. In vitro assays, on the other hand, often fail to reflect the complex multicellular and physicochemical context of the intact lung. In the present project, we will apply a unique microvasculature-on-chip platform that we have successfully developed in close collaboration with microphysiological model experts in Switzerland for the study of pericyte-endothelial cell interactions in pulmonary microvascular networks, and that we will employ here to study the effects of a targeted loss of pericytes on pulmonary microvascular network stability and the associated lung endothelial phenotype. Specifically, we aim to realize the following research aims:
Aim 1: To establish a system for targeted pericyte loss in pulmonary microvasculatures-on-a-chip. Pericyte loss will be induced by batrachotoxin, an activator of voltage-gated Na+ channels which are present in pericytes yet not in endothelial cells. As such, batrachotoxin should cause sustained membrane depolarization and ultimately, cell death only in pericytes. Induction of selective pericyte cell death will be assayed in cultured human lung pericytes and pulmonary microvascular endothelial cells, as well as in our pulmonary microvasculatures-on-a-chip platform.
Aim 2: To study the effects of targeted pericyte loss on microvascular structure in a pulmonary microvasculatures-on-a-chip platform. To this end, selective pericyte cell death will be induced by batrachotoxin in pulmonary microvasculatures-on-a-chip generated from cultured human lung pericytes and pulmonary microvascular endothelial cells, and effects on microvasculature network morphology and function will be studied by real-time imaging and digital image analysis.
Aim 3: To study the effects of targeted pericyte loss on the transcriptomic and functional phenotype of microvascular endothelial cells in a pulmonary microvasculatures-on-a-chip platform. Following targeted pericyte loss, endothelial cells will be retrieved from pulmonary microvasculatures-on-a-chip and undergo transcriptomic analysis by single cell RNA sequencing and phenotypic characterization.
References
- Bichsel CA, Hall SR, Schmid RA, Guenat OT, Geiser T. Primary Human Lung Pericytes Support and Stabilize In Vitro Perfusable Microvessels. Tissue Eng Part A. 2015; 21(15-16):2166-2176.
- Ferrari D, Sengupta A, Heo L, Pethö L, Michler J, Geiser T, […], Kuebler WM, Zeinali S, Guenat OT. Effects of biomechanical and biochemical stimuli on angio- and vasculogenesis in a complex microvasculature-on-chip. iScience. 2023; 26(3):106198.
- Zhang Q, Yaoita N, Tabuchi A, Liu S, Chen SH, Li Q, […], Verma S, Baker AH, Kuebler WM. Endothelial Heterogeneity in the Response to Autophagy Drives Small Vessel Muscularization in Pulmonary Hypertension. Circulation. 2024; 150(6):466-487.
- Bordenave J, Tu L, Berrebeh N, Thuillet R, Cumont A, Le Vely B, […], Humbert M, Huertas A, Guignabert C. Lineage tracing reveals the dynamic contribution of pericytes to the blood vessel remodeling in pulmonary hypertension. Arterioscler Thromb Vasc Biol. 2020; 40(3):766-782.
- Ricard N, Tu L, Le Hiress M, Huertas A, Phan C, Thuillet R, […], Dorfmüller P, Humbert M, Guignabert C. Increased pericyte coverage mediated by endothelial-derived fibroblast growth factor-2 and interleukin-6 is a source of smooth muscle-like cells in pulmonary hypertension. Circulation. 2014; 129(15):1586-1597.
- Yuan K, Liu Y, Zhang Y, Nathan A, Tian W, Yu J, […], Zamanian RT, Nicolls MR, de Jesus Perez VA. Mural Cell SDF1 Signaling Is Associated with the Pathogenesis of Pulmonary Arterial Hypertension. Am J Respir Cell Mol Biol. 2020; 62(6):747-759.
- Yuan K, Shamskhou EA, Orcholski ME, Nathan A, Reddy S, Honda H, […], Tian W, Nicolls MR, de Jesus Perez VA. Loss of Endothelium-Derived Wnt5a Is Associated With Reduced Pericyte Recruitment and Small Vessel Loss in Pulmonary Arterial Hypertension. Circulation. 2019; 139(14):1710-1724.
- Hariharan A, Weir N, Robertson C, He L, Betsholtz C, Longden TA. The Ion Channel and GPCR Toolkit of Brain Capillary Pericytes. Front Cell Neurosci. 2020; 14:601324.
- Lahiani A, Yavin E, Lazarovici P. The Molecular Basis of Toxins’ Interactions with Intracellular Signaling via Discrete Portals. Toxins. 2017; 9(3):107.
Uncovering molecular mechanisms leading to the onset of rheumatoid arthritis
Principal Investigator
Scientific interest within the context of the graduate college:
Rheumatoid arthritis (RA) represent a chronic and prevalent autoimmune disease characterized by inflammation and progredient joint destruction. Both anti-citrullinated protein antibodies (ACPA) and autoreactive ACPA-producing B cells play a key role in disease pathogenesis. However, the exact mechanisms leading to disease onset remain poorly understood. Notably, ACPA are not only present in RA patients, but can be also detected in a certain fraction of healthy individuals where they are indicating an increased risk of developing RA in the future. The proposed study thus aims to investigate the role of autoreactive ACPA-producing B cells in ACPA-positive healthy individuals and to understand the molecular mechanisms that eventually lead to the onset of inflammatory disease. By focusing on autoreactive B cells, we aim to uncover novel molecular insights into the pathogenesis of RA and identify potential therapeutic targets to prevent disease progression.
Project description:
We have collected a cohort of ACPA-positive healthy individuals as well as of ACPA-positive RA patents. The project aims to perform a molecular phenotyping of B cells in these patients where we seek to understand the behavior, characteristics and differences of autoreactive cells in ACPA-positive healthy individuals and RA patients. We plan to apply cutting-edge technology including bar-coded tetramers, mass cytometry and single-cell sequencing to characterize autoreactive ACPA-producing B cells. The overall aim is to understand the mechanisms that lead to onset of disease.
Aim 1: Examine the phenotypic and functional characteristics of autoreactive B cells in ACPA-positive individuals and RA patients using mass cytometry. We will use mass cytometry to perform a deep phenotyping of PBMCs of ACPA-positive individuals and RA patients where we will focus on activation markers and signaling pathway activity in response to ex vivo stimulation.
Aim 2: Examine the phenotypic and functional characteristics of autoreactive B cells in ACPA-positive individuals and RA patients using scRNAseq. Using scRNAseq and bar-coded tetramers, we will focus on the molecular phenotype of ACPA-producing autoreactive B cells in ACPA-positive individuals and RA patients.
Aim 3: Clinical Correlation. Access to clinical datasets will allow assessment of the correlation between B cell characteristics and clinical outcomes in ACPA-positive individuals.
References
- Schett G, Nagy G, Krönke G, Mielenz D. B-cell depletion in autoimmune diseases. Ann Rheum Dis. 2024; 83(11):1409-1420.
- Wilhelm A, Chambers D, Müller F, Bozec A, Grieshaber-Bouyer R, Winkler T, […], Mackensen A, Schett G, Krönke G. Selective CAR T cell-mediated B cell depletion suppresses IFN signature in SLE. JCI Insight. 2024; 9(12):e179433.
- Mackensen A, Müller F, Mougiakakos D, Böltz S, Wilhelm A, Aigner M, […], Winkler TH, Krönke G, Schett G. Anti-CD19 CAR T cell therapy for refractory systemic lupus erythematosus. Nat Med. 2022; 28(10):2124-2132.
- Scherer HU, Huizinga TWJ, Krönke G, Schett G, Toes REM. The B cell response to citrullinated antigens in the development of rheumatoid arthritis. Nat Rev Rheumatol. 2018; 14(3):157-169.
- Pfeifle R, Rothe T, Ipseiz N, Scherer HU, Culemann S, Harre U, […], Nimmerjahn F, Schett G, Krönke G. Regulation of autoantibody activity by the IL-23-TH17 axis determines the onset of autoimmune disease. Nat Immunol. 2017; 18(1):104-113.
Monitoring neuronal adaption to Toxoplasma gondii infection on the whole transcriptome level in the central and enteric nervous system
Principal Investigator

Prof. Dr. Ildiko Dunay
Scientific interest within the context of the graduate college:
Our lab investigates fundamental mechanisms of communication between the nervous and immune systems, two major sensory networks that continuously monitor tissue integrity and initiate protective responses upon disruption. While the immune system’s role in maintaining barrier function is increasingly understood, the contribution of neuronal signals to immune regulation remains largely unexplored. Recent work from the lab has revealed how neuropeptides such as neuromedin U and neurotransmitters like norepinephrine modulate innate lymphoid cells (ILCs), key tissue-resident immune cells at mucosal barriers. By integrating advanced genetic tools with state-of-the-art methods from immunology, neuroscience, and genomics, the lab aims to dissect the cellular and molecular circuits of neuro-immune interaction.
Project description:
Toxoplasma gondii, a ubiquitous intracellular parasite with a profound ability to infect virtually all warm-blooded animals, is responsible for a significant public health concern. In humans, infection can lead to toxoplasmosis, which, while often asymptomatic in healthy individuals, poses severe risks to immunocompromised patients and pregnant women.1 Moreover, emerging evidence suggests T. gondii’s potential triggering of various neurological disorders, including schizophrenia and other psychiatric conditions, underscoring the critical need to understand the parasite’s interaction with the nervous system.2 The central and enteric nervous systems are intricate networks that regulate numerous vital functions, from cognition and sensory processing in the central nervous system (CNS) to gastrointestinal motility and secretion in the enteric nervous system (ENS).3 T. gondii’s ability to invade and persist in these neural environments suggests significant adaptations by both the parasite and host neurons. Investigating these adaptations, particularly at the transcriptional level, provides insights into the mechanisms of pathogenesis, neuronal resilience, and potential neuropathology associated with infection.
We aim to monitor the transcriptional changes occurring in neurons during T. gondii infection. To this end, we have already established the respective lines and protocols for bulk sequencing of purified RNA from sort-purified GFP+ neuronal nuclei from the T. gondii-infected brain and gut tissue of the Snap25Cre/+ x INTACT system, in which the GFP expression is only activated in neurons. To monitor alterations in enteric neurons’ composition and transcriptional changes in subsets of enteric neurons, we will perform RNA sequencing from brains and gut tissue. To this end, we will separately sort-purify GFP+ nuclei from Snap25Cre/+ x INTACT system to obtain highly pure neuronal nuclei. These neuronal cell nuclei will be analyzed using the RNA-sequencing platform, which we have already implemented for single-nuclei sequencing. Together, these experiments will identify genes in neurons regulated in the context of T. gondii infection.
Based on our data detecting a type II interferon signature in the ENS in various inflammation models, we will perform bulk RNA sequencing of the ENS and CNS, in which we have deleted the Ifngr1 specifically in neurons by the Snap25Cre/+ x Ifngr1fl/fl INTACT system and which allow for sort-purification based on the nuclear GFP signal. This experimental setup will allow us to study the potentially protective gene expression signature in neurons induced by IFN-γ. Furthermore, these experiments will enable us measure if neuron-specific deletion of the IFN-γ receptor results in increased T.gondii burden in various organs. Therefore, these experiments will reveal how IFN-γ-signaling in neurons contributes to anti-toxoplasma immunity.
Taken together, these datasets will yield correlations of differentially expressed genes of ENS and CNS neurons and their relative abundances, exposing disease-relevant signaling circuits triggered in neurons. This global perspective is essential for uncovering the molecular underpinnings of T. gondii’s impact on neuronal function and survival, which could lead to the development of targeted therapies and interventions.1,2
References
- Dunay IR, Gajurel K, Dhakal R, Liesenfeld O, Montoya JG. Treatment of Toxoplasmosis: Historical Perspective, Animal Models, and Current Clinical Practice. Clin Microbiol Rev. 2018; 31(4):e00057-17.
- Matta SK, Rinkenberger N, Dunay IR, Sibley LD. Toxoplasma gondii infection and its implications within the central nervous system. Nat Rev Microbiol. 2021; 19(7):467-480.
- Jakob MO, Kofoed-Branzk M, Deshpande D, Murugan S, Klose CSN. An Integrated View on Neuronal Subsets in the Peripheral Nervous System and Their Role in Immunoregulation. Front Immunol. 2021; 12:679055.
Multi-modal spatial characterization of fibrovascular skin niches in health and disease
Prinicipal Investigator

Dr. Elise Siegert
Scientific interest within the context of the graduate college:
We are interested in the interaction of endothelial cells and their mesenchymal neighboring cells with immune cells in perivascular tissue areas called fibrovascular niches. These represent crucial strategic sites for resident immune cells regulating tissue homeostasis and maintain structural and functional integrity of blood vessels in health and disease. Factors disturbing structural and functional integrity of fibrovascular niches are associated with pre-mature aging and irreversible organ damage. However, the cellular interplay and molecular basis of those processes are not well understood.
Project description:
Located at the interface between the blood circulation and tissue, fibrovascular niches are strategically positioned to transmit information about systemic conditions to tissues (Pascual-Reguant et al., 2024). This exposition to systemic changes makes them a starting point for certain pathologies, for example tissue fibrosis, as we recently demonstrated (Mothes et al., 2023). Advanced imaging technologies allow us to characterize tissue composition at a cellular and molecular level in various modalities, detecting protein as well as RNA in tissue sections. Using those technologies, we aim to better understand fibrovascular niche functions in vascular health, physiological neovascularization and wound healing as well as in autoimmune rheumatic diseases.
We hypothesize that the spatial organization of various cellular compartments (immune cells, endothelial and mesenchymal cells) in fibrovascular niches is a crucial component of vascular integrity and impacts on functions of the skin in health and disease. We know from previous studies on wound healing that the interaction between macrophages and fibroblasts is a key component in determining the differential functional states of wound healing (Adler et al., 2020). Their reciprocal activation is crucial for scar maturation and results in an upregulation of genes related to epithelial-to-mesenchymal transition and angiogenesis in fibroblasts (Setten et al., 2022).
We will compare cellular composition, cell-cell interactions and molecular pathways under physiological conditions, in physiological wound healing and pathological scaring in the context of the fibrotic autoimmune disease systemic sclerosis; the latter is characterized by diffuse fibrosis and vascular anomalies in skin, joints and organs such as gastrointestinal tract, heart, lungs and kidneys. Whilst disease mechanisms at the tissue level in systemic sclerosis are still incompletely understood, B cell depletion has recently led to many promising therapeutic outcomes (Auth et al., 2025; Siegert et al., 2025).
Aim 1: Determining role of the different cellular components in fibrovascular niche in physiological homeostasis and physiological wound healing, comparison to pathological states such as early systemic sclerosis. We will collect punch biopsy skin samples from healthy controls and from fibrotic skin from systemic sclerosis patients (disease duration of less than two years as defined by onset of first non-Raynaud symptom). We will use spatial transcriptomics for molecular tissue profiling at sub-cellular resolution and map it to the cellular and extra-cellular structures identified using multi-epitope ligand cartography. This will allow is to spatially map cellular and molecular changes occurring during pathologies in fibrovascular niches and identify potential new therapeutic targets.
Aim 2: Identifying cellular changes in the fibrovascular niche following plasma cell depleting therapy with teclistamab, a bispecific antibody against BCMA / CD3, in systemic sclerosis. Our preliminary data show that treatment with subcutaneous teclistamab injections is highly effective in reducing skin fibrosis, resolving inflammation and promoting neoangiogenesis in systemic sclerosis skin (Siegert et al., 2025). We will collect 4 mm punch biopsies skin samples from patients undergoing treatment with teclistamab at baseline and at week 12 after treatment. We will perform untargeted spatial transcriptomics to identify target genes and pathways which we will validate by multiplex immunofluorescence histology. This approach will enable us to identify how the cellular alterations of the fibrovascular niche caused by the B lineage-depleting therapy translate into structural and functional alterations of the vasculature and their capacity to regenerate.
References
- Adler M, Mayo A, Zhou X, Franklin RA, Meizlish ML, Medzhitov R, Kallenberger SM, Alon U. Principles of Cell Circuits for Tissue Repair and Fibrosis. iScience 2020; 23(2):100841.
- Auth J, Müller F, Völkl S, Bayerl N, Distler JHW, Tur C, […], Mackensen A, Schett G, Bergmann C. CD19-targeting CAR T-cell therapy in patients with diffuse systemic sclerosis: a case series. Lancet Rheumatol. 2025; 7(2):e83-e93.
- Mothes RA, Pascual-Reguant R, Koehler J, Liebeskind J, Liebheit A, Bauherr S, […], Niesner RA, Radbruch H, Hauser AE. Distinct tissue niches direct lung immunopathology via CCL18 and CCL21 in severe COVID-19. Nat Commun. 2023; 14(1):791.
- Pascual-Reguant A, Kroh S, Hauser AE. Tissue niches and immunopathology through the lens of spatial tissue profiling techniques. Eur J Immunol. 2024; 54(2):e2350484.
- Setten E, Castagna A, Nava-Sedeño JM, Weber J, Carriero R, Reppas A, […], Hatzikirou H, Feuerhake F, Locati M. Understanding fibrosis pathogenesis via modeling macrophage-fibroblast interplay in immune-metabolic context. Nat Commun. 2022; 13(1):6499.
- Siegert E, Biesen R, Dzamukova M, Furth C, Probst M, Doellinger F, […], Krönke J, Krönke G, Alexander T. Teclistamab in relapsed systemic sclerosis after autologous haematopoietic stem cell transplantation. Ann Rheum Dis. 2025; 84(4):653-656.
The influence of lipidome alterations on dendritic cell functionality in fatty liver (MASLD)
Principal Investigator

Dr. Leke Wiering
Prof. Dr. Frank Tacke
Scientific interest within the context of the graduate college:
Our lab studies the role of immune cells and inflammatory processes in the liver. During homeostasis the liver plays an important role in adaptation to environmental influences as it is constantly exposed to antigens from the gastrointestinal system, and plays a critical role in maintaining a balance between tolerance to harmless antigens (eg. food proteins or commensal bacteria) and control of pathogens.1 When this balance is disrupted (termed maladaptation) the resulting immune-mediated changes can lead to chronic liver diseases and ultimately cancer.2 Infiltration and activation of immune cells play an important role during the development of liver diseases, but the exact molecular and cellular mechanisms leading to the development of liver inflammation have not been fully elucidated until now. We are exploring the inflammatory processes during acute liver failure, metabolic dysfunction-associated steatotic liver disease and steatohepatitis (MASLD/MASH), liver cirrhosis and liver cancer in order to develop new diagnostic and therapeutic strategies. Furthermore, a better understanding of how both pro- and anti-inflammatory pathways can disrupt the homeostatic processes of a healthy liver is critical for the prevention of liver diseases in the first place.
Project description:
Metabolic dysfunction-associated steatotic liver disease (MASLD) is the most common chronic liver disease worldwide, affecting a staggering 30% of the general population, and its prevalence is expected to rise further with an aging and more obese world population. MASLD is characterized by persistent inflammation and subsequent liver fibrosis, which can progress to steatohepatitis, cirrhosis and cancer. Dendritic cells (DC) have been shown to accumulate in chronic liver diseases, but their exact role in liver inflammation and fibrosis is complex and incompletely understood. In the context of MASLD/MASH, specific DC subsets correlate with disease severity in patients and drive liver pathology in mouse models.3 Furthermore, it has been shown that the lipid content of DC can significantly influence their functionality, with “lipid-high” DC displaying a proinflammatory phenotype while “lipid-low” DC seem to be rather tolerogenic.4 In this project, we would like to study how changes in the hepatic lipidome influence infiltration, differentiation and function of DCs and how this could be targeted to prevent or reverse the development of MASLD. For this purpose, tissue samples from both patients and mice with MASLD/MASH will be analyzed with state-of-the-art single-cell technologies, such as full spectrum flow cytometry,5 imaging mass cytometry and mass spectrometry imaging.6 This will be complemented by functional in vitro and ex vivo experiments using 2D and 3D cell culture systems.
Aim 1: Characterize the relationship between the hepatic lipidome and dendritic cell subsets in MASLD. Human and murine liver samples will be analyzed by imaging to mass cytometry to determine infiltration patterns of DC subsets and mass spectrometry imaging to characterize the hepatic lipid environment(s). Correlation of the results will determine which lipid species are associated with DC infiltration and distribution in the tissue.
Aim 2: Understand the influence of lipid species on dendritic cell differentiation and maturation in vitro. In vitro culture of primary human and murine dendritic cells with lipid species identified in Aim 1 will be used to analyze their influence on DC differentiation, activation and maturation. Readouts will include full spectrum flow cytometry, cytokine measurements as well as functional assays (e.g. antigen presentation).
Aim 3: Modulate dendritic cell functionality in vivo. Human and mouse hepatic organoids will be used to test how DC function can be targeted to prevent differentiation into pathogenic phenotypes, e.g. through inhibition of uptake of certain lipid species and/or reprogramming of intracellular DC lipid metabolism.
References
- Kubes P and Jenne C. Immune Responses in the Liver. Annu Rev Immunol. 2018; 36:247-277.
- Hammerich L and Tacke F. Hepatic inflammatory responses in liver fibrosis. Nat Rev Gastroenterol Hepatol. 2023; 20(10):633-646.
- Deczkowska A, David E, Ramadori P, Pfister D, Safran M, Li B, […], Heikenwälder M, Elinav E, Amit I. XCR1(+) type 1 conventional dendritic cells drive liver pathology in non-alcoholic steatohepatitis. Nat Med. 2021; 27(6):1043-1054.
- Ibrahim J, Nguyen AH, Rehman A, Ochi A, Jamal M, Graffeo CS, […], Acehan D, Frey AB, Miller G. Dendritic cell populations with different concentrations of lipid regulate tolerance and immunity in mouse and human liver. Gastroenterology. 2012; 143(4):1061-1072.
- Hammerich L, Shevchenko Y, Knorr J, Werner W, Bruneau A, Tacke F. Resolving 31 colors on a standard 3-laser full spectrum flow cytometer for immune monitoring of human blood samples. Cytometry B Clin Cytom. 2023; 104(5):367-373.
- Goossens P, Lu C, Cao J, Gijbels MJ, Karel JMH, Wijnands E, […], Donners MMPC, Heeren RMA, Biessen EAL. Integrating multiplex immunofluorescent and mass spectrometry imaging to map myeloid heterogeneity in its metabolic and cellular context. Cell Metab. 2022; 34(8):1214-1225.e6.
Molecular and clinical analysis of the most frequent European Alport syndrome variant
Principal Investigator

Dr. Ria Schönauer
Scientific interest within the context of the graduate college:
Not Everything Is “Genetic”, but Genes Are Involved in Everything (adapted from Kenneth M. Weiss). Our group is interested in identification and investigation of genetic, clinical, and environmental factors determining onset of chronic kidney disease (CKD) and kidney survival. We make use of next-generation sequencing techniques and deep-phenotyping to identify genetic variants that are predictive for disease progression or convey protection from organ failure. We functionally evaluate identified germline variants in vitro in order to understand underlying molecular mechanisms leading to CKD on the one hand or protecting from kidney failure on the other. By doing so, we aim at defining and targeting molecular switches responsible for health maintenance and disease alleviation.
Project description:
Alport syndrome (AS) is one of the most common genetic kidney diseases. On the renal level, AS is characterized by proteinuria, hematuria, and progressive kidney function decline over lifetime. Beyond the kidney, affected individuals may suffer from sensorineural deafness and eye involvement, such as retinopathy, cataract and lenticonus. AS is caused by germline mutations in three disease genes: COL4A3, COL4A4, and COL4A5. While mutations in the latter lead to X-linked AS with a male preponderance, defects in COL4A3 and COL4A4 result in the autosomal forms, either with a dominant or recessive inheritance pattern. There is one single gene variant in COL4A5 accounting for about 16% of all AS-cases in our datasets: COL4A5-Gly624Asp.1 This variant constitutes a founder mutation originating from Eastern Europe. Interestingly, we and others found that it is associated with a significantly milder course of disease: median age at kidney failure between 20-30 years later than in carriers of other COL4A5 missense mutations.1,2 However, the molecular effect of the most common disease causing COL4A5 variant has not been investigated to date and it is unknown why it is somehow less damaging than other protein variants. On the other hand, we observe a phenomenon called intra- and inter-familial variability among carriers of COL4A5-Gly624Asp, leaving the possibility of environmental and epigenetic modification. To address this question, we propose the following two specific aims and work packets (WP):
Aim 1 / WP1: Clinical cohort expansion and functional comparison of COL4A5-Gly624Asp compared to other COL4A5-missense variants. With the help of the European Reference Network (ERKNet), we aim to identify and clinically characterize additional patients with COL4A5-Gly624Asp. Functionally, we would like to run in vitro assays in overexpression systems (such as Western Blotting /Co-IP, by use of HEK293T) for systematic comparison of COL4A5-Gly624Asp with other Gly- and non-Gly missense variants. Thereby, we aim to see differences in residual protein function, stability, and protein interaction compared to other COL4A4/A3/A5 mutant and wildtype constructs.
Aim 2 / WP2: Epigenetic comparison of monozygotic twins with the COL4A5-Gly624Asp variant. DNA methylation is an epigenetic modification that plays an important role in regulating gene expression. Within our in-house AS-cohort (n> 200), we have a unique opportunity to investigate this research question by examining divergent disease progression in monozygotic twins carrying the identical COL4A5 Gly624Asp variant, yet exhibiting markedly different disease severity (CKD stage G3a in one twin versus CKD stage G5 in the other, both at 32 years of age).We would like to investigate their existing kidney biopsy material (kidney tissue) for methylome-wide differences potentially explaining discrepancies in clinical course (in collaboration with Dr. rer. nat. Antje Richter, expert on epigenetics at the Justus-Liebig University in Giessen).
References
- Krüger BM, Jens A, Neuhaus A, Ćomić J, Berutti R, de Fallois J, […], Schrezenmeier EV, Hoefele J, Halbritter J. COL4A5-p.Gly624Asp is the Predominant Variant in Europe Associated With a Mild Alport Syndrome Phenotype. Kidney Int Rep. 2025; 10(5):1372-1383.
- Żurowska AM, Bielska O, Daca-Roszak P, Jankowski M, Szczepańska M, Roszkowska-Bjanid D, […], Kuleszo D, Ziętkiewicz E, Lipska-Ziętkiewicz BS. Mild X-linked Alport syndrome due to the COL4A5 G624D variant originating in the Middle Ages is predominant in Central/East Europe and causes kidney failure in midlife. Kidney Int. 2021; 99(6):1451-1458.



